SS 2025: Drivers of plant and fungal diversity: from genomes to biomes
List of topics is still in progress
Phylogenetic relationships of Mexican Tillandsia (Bromeliaceae)
This project will explore the phylogenetic relationships within Tillandsia subgenus Tillandsia, a diverse bromeliad adaptive radiation. The student will gain hands-on experience in molecular systematics by contributing to DNA extractions, Illumina library preparations, and phylogenomic analyses. The project will produce new data on 17 Mexican Tillandsia species, which combined with pre-existing data will help understand evolutionary history and provide insights into diversification patterns within this iconic bromeliad group.
Supervisors: Ovidiu Paun & Michael Barfuss

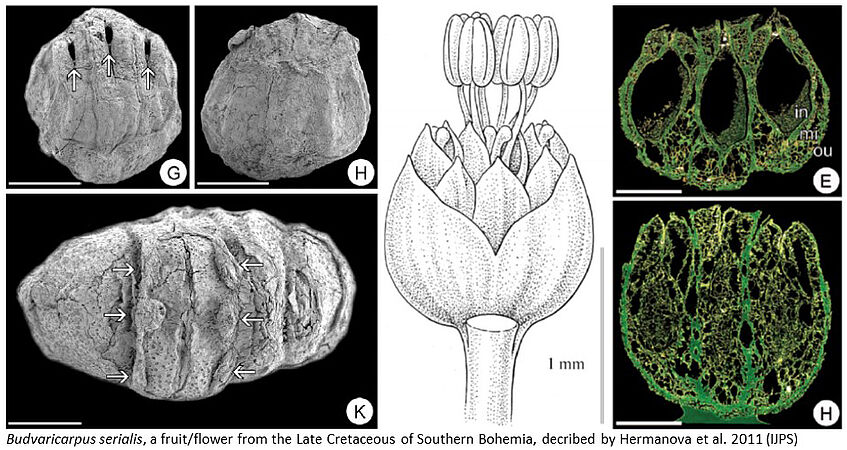
Three-dimensionally preserved fossil fruits from the Cretaceous of Southern Bohemia
The Cretaceous fossil record is crucial for our understanding of angiosperm diversification. Together with paleobotanists from Prague, we have recently collected Late Cretaceous sediments (ca. 90 million year old) from Southern Bohemia containing numerous charcoalified angiosperm fossils (mostly tiny fruits and seed). The goals of this specific research project will be (1) to extract fossils by sieving a sediment sample, (2) to sort and identify potentially interesting fossil fruits, and (3) to morphologically analyse and describe a small number of selected fossils. Methods will involve Scanning Electron Microscopy and High Resolution X-Ray Computed Tomography. The project has the potential to be extended into a master thesis.
Supervisors: Jürg Schönenberger & Maria von Balthazar
Fossil flowers with in-situ pollen from the Eocene of Germany
Compressed flowers from the Eocene of Germany will be described and their in-situ pollen investigated with light microscopy (LM), scanning electron microscopy (SEM), and transmission electron microscopy (TEM). The fossil flowers and the pollen grains will be compared to flowers and pollen from potential modern analogues (PMAs). Detailed descriptions based on morphological and ultrastructural features of the pollen will be used to affiliate the flowers and the in-situ pollen to extinct or extant angiosperm taxa. The paleoecological aspects of the fossil plants will be evaluated based on ecological preferences of PMAs.
Supervisors: Friðgeir Grímsson & Silvia Ulrich

Comparative analyses of repetitive DNA profiles of the major bryophyte lineages
Repetitive DNA is an important and often dominant component of most plant genomes. Most of the genomes characterized so far belong to angiosperms, and very little is known about the genomic landscape of repetitive DNA families in bryophytes. This projects aims to analyze the repeats of the representatives of three major lineages of bryophytes (hornworts, liverworts and mosses) using NGS (Illumina) genome skimming data. The sequences will be clustered using the RepeatExporer pipeline and the repetitive elements populating the genomes will be annotated and characterized. These will subsequently be used for comparative analyses. The project will provide first comprehensive insight into the repeat families in bryophytes. Novel satellite DNAs will be isolated for further cytogenetic analyses.
Supervisor: Hanna Schneeweiss

Environmental stress at the cellular level
This research project is focussed on osmotic stress of plant cells. In order to visualize the shape of cells with and without turgor, different specific dyes will be tested. The anatomy of leaves in seed plants (cross sections) or in mosses will be studied using microscopy, cell biological methods or microtome sections.
Supervisor: Ingeborg Lang
Vigorous Primula auricula under harsh conditions – do specialized metabolites (SPM) help out?
We use the alpine Primula auricula as a model plant for chemical diversification studies. This plant can grow even under harsh environmental conditions, which may be due to the exceptional production of a mostly mealy excretion, referred to as farina, being produced by glandular trichomes of aerial parts. Exudates contain enormous amounts of unsubstituted flavone, indicative of a functional role for the plant. So far, the influence of abiotic factors on both farina and flavone production was studied by our group. Now we want to extend our research towards biotic interactions, for a better understanding of flavone accumulation in an ecological context. However, flavonoids are not the only constituents. Similarly, other parts of the plants may also play a role for a vigorous growth of P. auricula in its natural habitat. Thus, rhizomes – the overwintering belowground stems – came into focus recently. We want to extend our analyses also to this part of the plant, both in respect to chemical composition and ecological factors of SPM accumulation. General analytical procedures applied comprise various chromatographic techniques, both for comparative analyses and for isolation of interesting compounds. As there are hardly any competitors growing near P. auricula in the field, flavone and other chemicals released may have allelopathic effects that need to be assessed in this project - just one more piece to complete our puzzled view on this species.
Supervisors: Johann Schinnerl & Karin Vetschera

Evolution of repetitive DNAs in cultivated and wild chile peppers (Capsicum, Solanaceae)
Chiles (Capsicum spp., Solanaceae) are well-known vegetables and spices consumed worldwide. Most cultivated varieties belong to three species, but the genus includes over 40 wild species native to Central-South America. Repetitive DNA is an important and often dominant component of most plant genomes. This project aims to analyse the abundance, chromosomal distribution and the evolutionary dynamics of newly identified selected satellite DNAs (tandem repeats) in cultivated and wild pepper species. The repeats will be mapped in chromosomes via fluorescence in situ hybridization (FISH) and the data analysed using fluorescence microscopy. The data will be interpreted in a phylogenetic context and implemented with basic NGS sequence data analyses (user friendly) to provide new insights into the evolution of chile genomes. Mapping of the novel satellite DNAs will also provide chromosomal markers to aid individual chromosome identification.
Supervisor: Hanna Schneeweiss

Evolutionary history of Hawaiian Diospyros (Ebeneaceae)
This student project is part of a larger research initiative investigating the drivers of a plant radiation in New Caledonia, a globally recognized biodiversity hotspot. The focus of this project will be on a Hawaiian species pair, which forms the sister clade of the radiated New Caledonian Diospyros lineage. Depending on the student’s interest, project could involve genomic analyses to infer species boundarie, reconstruct historical changes in population size, investigate patterns of gene flow, of explore genomic landscape evolution. This research will provide valuable insights into the evolutionary processes shaping island plant diversity and contribute to a broader understanding of adaptive radiation and speciation.
Supervisor: Ovidiu Paun

morphological variability of the Tinder Mushooms in East Austria
You will study host range, host specificity, variability of discriminating morphological basidioma characters and do barcoding of the Fomes fomentarius alliance in East Austria. You will work with a 3-D-microscope for pore measurements, doing DNA lab work and phylogenetic analysis. The Fomes fomentarius agg. has at least two taxonomic entities in Austria, boundaries and host ranges are still unclear.
Supervisors: Irmgard Greilhuber & Michael Barfuss

Origin and evolution of polyploid species in Melampodium (Asteraceae)
The origin of polyploids and its impact on the dynamics of plant genome evolution is of major interest in evolutionary botany. In this project RAD-seq data will be used to establish the phylogenetic framework in which the recurrent and nested origin of polyploids (tetraploids and hexaploids) will be tested. Furthermore, these data will form the basis for identifying the geographic origin and its possible ecological correlates of the polyploids.
Supervisors: Hanna Schneeweiss & Gerald M. Schneeweiss
Assessing ancient range shifts from molecular data: historical biogeography of Phyteuma (Campanulaceae)
Plants responded to Pleistocene climate oscillations by range shifts, which may result in lineage diversification and speciation. In the absence of fossil evidence (lacking for the vast majority of herbaceous species), past range shifts have to be inferred from genetic data. During a previous student's project, it has been found that within Phyteuma spicatum (Campanulaceae), a species widespread in temperate Europe, a few populations from the central Alps and northern Apennines possess chloroplast haplotypes very different from those found in the other populations. These haplotypes could be remnants of a more widespread ancestor of the P. spicatum group suggested by the sister relationship of the geographically disjoint taxa P. pyrenaicum, restricted to the Iberian Peninsula, and P. vagneri, endemic to the Carpathians. To test this hypothesis, the existing data set of plastid sequences from P. spicatum will be extended by data from closely related species (P. gallicum, P. nigrum, P. pyranaicum, P. vagneri) obtained via Sanger DNA sequencing.
Supervisors: Michael Barfuss & Gerald M. Schneeweiss

