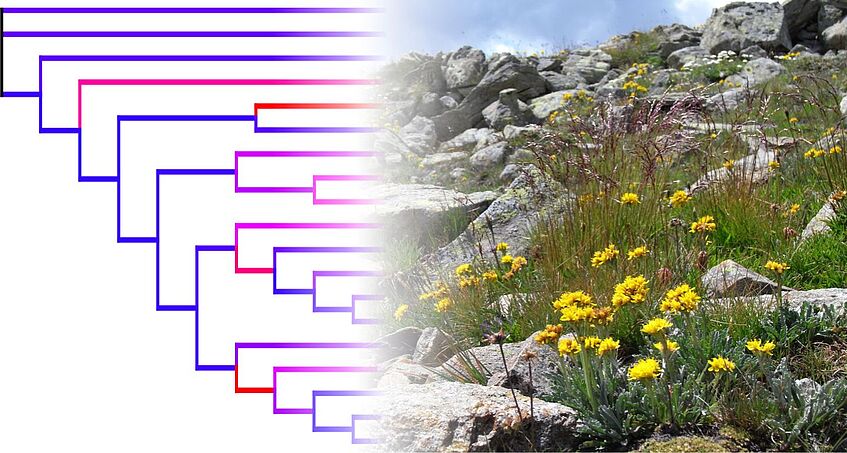
SS 2019: Drivers of plant and fungal diversity: from genomes to biomes
Gene expression plasticity along altitude in Heliosperma
Plant distribution along altitude offers a suitable model to understand plant responses to climatic changes. The ecological parameters that differ along altitude include average temperatures, distinct day-night temperature amplitudes, light and moisture regimes, together with a different biotic environment. Plant responses to such wide-ranging ecological difference will comprise plastic, epigenetic and genetic components, but their extent and ecological relevance are still not well understood. This project will investigate gene expression (RNAseq) plasticity across reciprocal transplantations between mountain and alpine localities of Heliosperma pusillum (Caryophyllaceae) in the Alps (see photos).
Supervisor: Ovidiu Paun

Meta-genomic analysis of the Populus alba (white poplar) microbiome
Whole genome sequencing of plant material often includes DNA sequences that cannot be mapped to the reference genome. This project will deal with mining whole genome sequence data from poplar forests across Europe for bacterial and fungal diversity.
Supervisors: Jaqueline Hess & Christian Lexer
Barcoding the genus Tricholomopsis in Austria
You will generate ITS sequence data and thus contribute to the big ABOL-project. How many species does the small genus Tricholomopsis actually include? Does Tricholomopsis flammula merit species level? Are there cryptic entities in T. rutilans and T. decora? Your source of material will be the fungarium of Vienna University.
Supervisors: Irmgard Greilhuber & Michael Barfuss
Optimized documentation and photography of bryophyte species
The aim of this project is to document features of bryophytes that are relevant for species identification. To this end, from up to five species macroscopic and light microscopic documentation will be made. The images will be optimized for the use in textbooks.
Supervisor: Ingeborg Lang

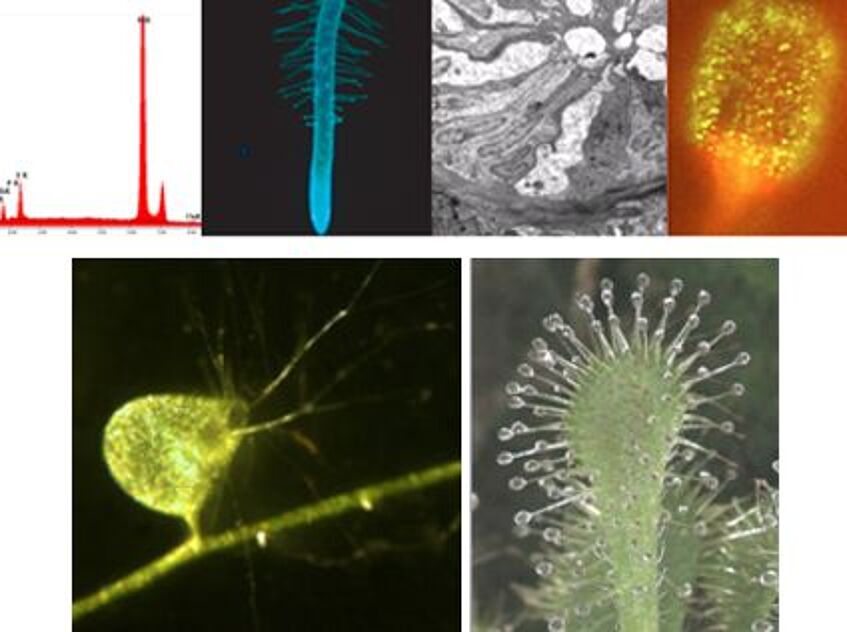
Structure and function of carnivorous plants
Carnivorous plants supplement their mineral nutrition by absorbing different elements from animals. For this aim, they have developed trap organs that carry glandular cells producing trapping mucilage and digestive enzymes. Also nutrient uptake often occurs via gland cells. Main topics include morphology and anatomy of the traps, structure of glandular systems, production of attractants and signaling, production of digestive enzymes, uptake of digested proteins, and movement of leaves upon signals from animals.
Supervisor: Irene Lichtscheidl
Transcriptomic drivers of the ecological divergence between sibling allopolyploids marsh orchids
Whole genome doubling and hybridization profoundly shaped plant genome evolution. To be successful, first generations polyploids must quickly adjust their genome and function, thereby altering their ecological properties and adaptive success, as a function of their environment. This project will investigate in detail the molecular basis of the eco-physiological divergence between two orchid tetraploids (Dactylorhiza), focusing for example on preferences for distinct soil types or different photosynthetic characteristics.
Supervisor: Ovidiu Paun

A comparison between PAM‐fluorescence and primary productivity
Pulse-Amplitude Modulation (PAM) is a technique permitting chlorophyll fluorescence to be measured quickly and in a non-invasive manner. This project will address the question whether the PAM-fluorescence technique can be used to provide reliable estimations of oxgyen production in microalgae.
Supervisor: Michael Schagerl
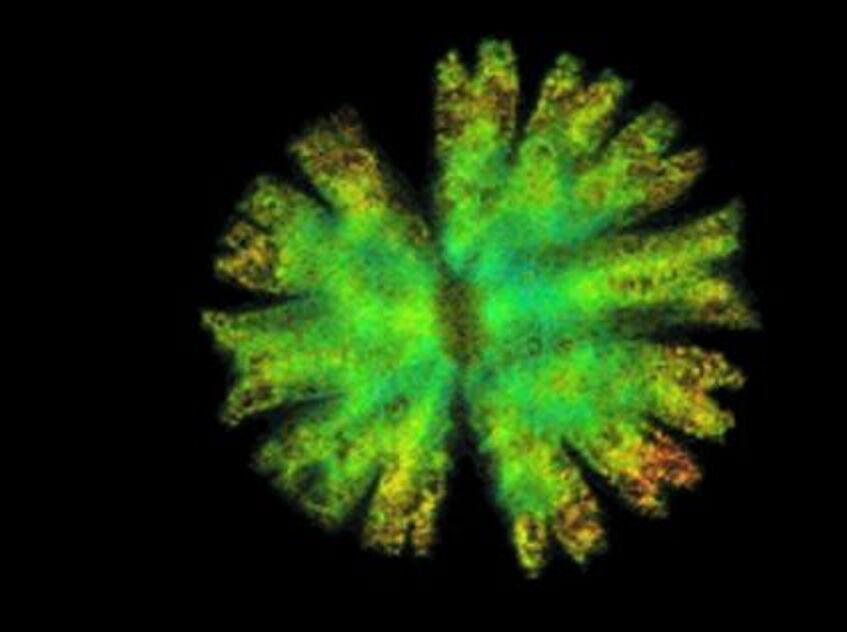
Phylogenetics and biogeography of Nicotiana section Suaveolentes
Nicotiana section Suaveolentes has a whole genome doubling in its ancestry and it is widespread in Australia, ranging from a few species in mesic areas to the great majority in semi-arid and arid regions. Hypotheses exist that the group first occupied the wetter N and E parts of Australia where all species with higher chromosome numbers occur. From there lineages may have colonized and adapted stepwise to increasingly arid areas, in parallel to chromosomal condensation and diploidization. With genomic data (RADseq) and phylogenomic methods this project will aim to test this hypothesis. (Photo: M. Christenhusz)
Supervisor: Ovidiu Paun & Mary Rosabelle Samuel

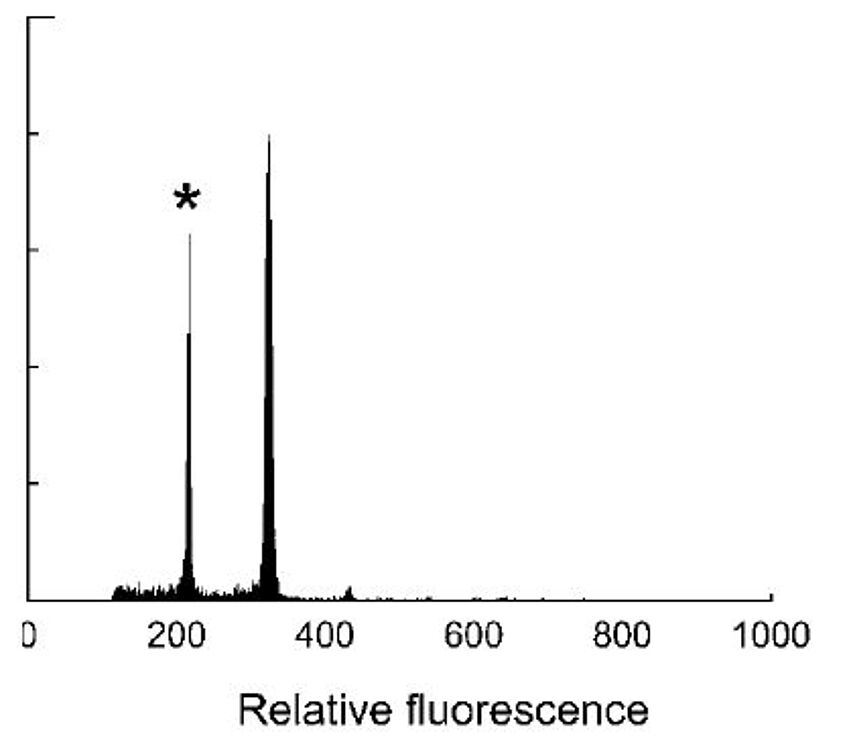
Cryptic diversity in the Austrian flora
Closely related species often differ in their ploidy level (diploid versus polyploid), buts they might be difficult to tell apart morphologically. Accordingly, their status (e.g., the extent of their distribution range in Austria) is insufficiently known. In the frame of this project, ploidy determination using flow-cytometry, DNA barcoding and, optionally, morphometric analyses will be used in selected plant groups.
Supervisors: Hanna Schneeweiss & Gerald M. Schneeweiss
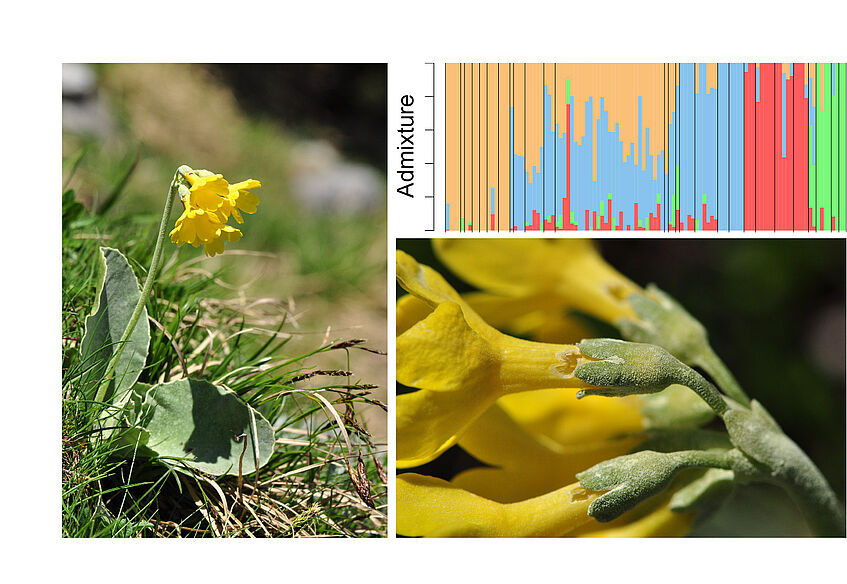
Evolution of Primula auricula complex in eastern Alps
The phenotypic diversity that is observable along an altitudinal gradient on Schneeberg for Primula auricula suggests a possible differentiation into subspecies or ecotypes. This project will apply population genomic (RADseq) to complement phytochemical analyses in order to investigate in detail the differentiation of populations along this altitudinal gradient. (Photos: C. Gilli)
Supervisor: Ovidiu Paun
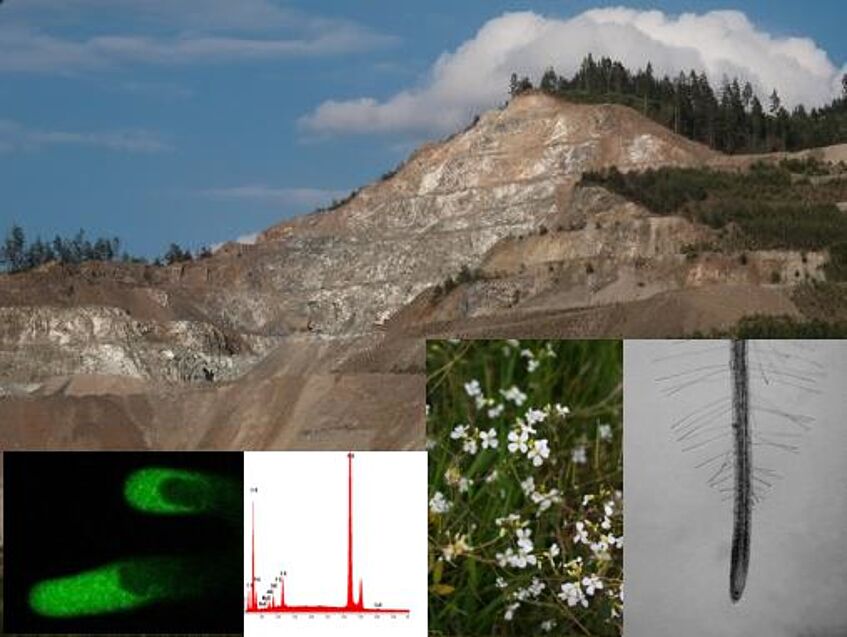
Structure and function of heavy metal tolerant plants
Many heavy metals are important micro-nutrients, but all become toxic if they are in too high concentration. Some plants, however, have developed strategies to deal with this difficult environmental situation. Topics offered include different strategies (tolerant plants, metal accumulators, excluders) as well as interactions of plants with soil microbes.
Supervisor: Irene Lichtscheidl
Evolution into alpine habitats
Alpine habitats are disproportionally species-rich given the surface area mountain areas occupy. A major evolutionary-macroecological question is whether lineages possessing traits rendering them ecologically pre-adapted are more successful (i.e., diverse) in alpine habitats than others.
Supervisor: Gerald M. Schneeweiss